Component | AP231-21-V4 | AP231-22-V4 | AP231-23-V4 |
| TransStart® FastPfu Fly DNA Polymerase | 250 U×1 | 500 U×1 | 500 U×6 |
| 2×TransStart® FastPfu Fly Reaction Mix | 1 ml×3 | 1.2 ml×5 | 1.2 ml×30 |
| 50 mM MgSO4 | 200 μl×1 | 400 μl×1 | 1 ml×1 |
| 6×DNA Loading Buffer | 500 μl×1 | 1 ml×1 | 1 ml×2 |
| PCR Stimulant | 200 μl×1 | 400 μl×1 | 1 ml×1 |
TransStart® FastPfu Fly DNA Polymerase
Cat# AP231-23-V4
Size : 6×500units
Brand : TransGen Biotech
TransStart® FastPfu Fly DNA Polymerase
Catalog Number: AP231-21-V4
Price:Please Inquiry First
Product Details
TransStart® FastPfu Fly DNA Polymerase is a hot start, ultra high-fidelity DNA Polymerase used for fast PCR. Compared with TransStart® FastPfu DNA Polymerase, TransStart®FastPfu Fly DNA Polymerase has higher extension rate (< 5 kb fragments can achieve 12 kb/min extreme amplification, > 5 kb fragments can achieve 6 kb/min high-speed amplification), featuring higher amplification efficiency, higher yield, higher fidelity and higher specificity. 2x TransStart® FastPfu Fly Reaction Mix already contains dNTPs. When DNA is amplified, just add template, primer, water and TransStart® FastPfu Fly DNA Polymerase. The Reaction can be carried out with the concentration of Reaction Mix being l x. PCR products are blunt end and can be cloned directly into pEASY®-Blunt series of vectors.
• Offers 108-fold fidelity as compared to EasyTaq®DNA Polymerase.
• PCR products can be directly cloned into pEASY®-Blunt vectors.
• Amplification of genomic DNA fragment up to 15 kb.
• Amplification of plasmid DNA fragment up to 20 kb.
• Hot start, high specificity.
• High amplification efficiency.
• Fast and ultra high-fidelity.
• High sensitivity.
• High yield.
• Complex templates, GC/AT-rich templates.
• Ultra high-fidelity and fast PCR, blunt end cloning, site-directed mutagenesis.
• Long fragment amplification.
-20°C for two years
Dry ice (-70°C)
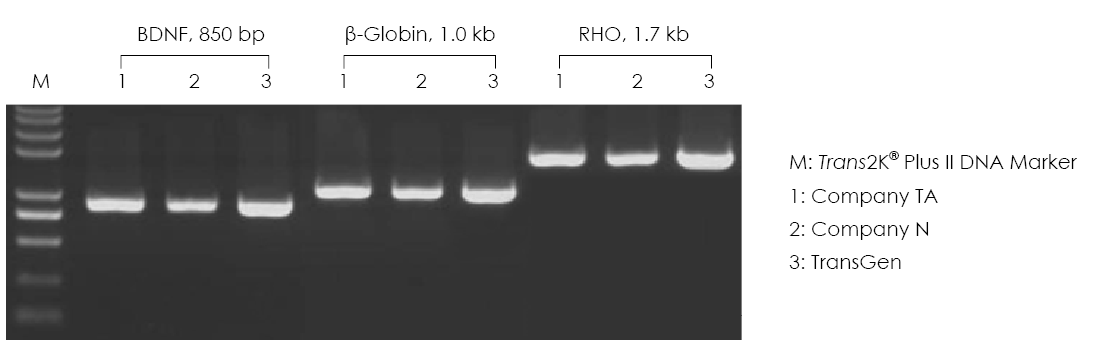
Using products from TransGen, Company TA, and Company N, amplify different genes by using human gDNA as templates, and analyze the amplification effect through 1.0% agarose gel electrophoresis.
1 Zhang A, Shan T, Sun Y, et al. Directed evolution rice genes with randomly multiplexed sgRNAs assembly of base editors[J]. Plant Biotechnology Journal, 2023.(IF 13.80)
2 Xu Y, Zhu T F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs[J]. Science, 2022.(IF 63.714)
3 Wang D, Yan F, Wu P, et al. Global profiling of regulatory elements in the histone benzoylation pathway[J]. Nature Communications, 2022.(IF 14.919)
4 Niu L, Shen W, Shi Z, et al. Three-dimensional folding dynamics of the Xenopus tropicalis genome[J]. Nature Genetics, 2021.(IF 38.33)
5 Jin S, Fei H, Zhu Z, et al. Rationally designed APOBEC3B cytosine base editors with improved specificity[J]. Molecular cell, 2020.(IF 15.58)
6 Chen J, Ou Y, Yang Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing[J]. Nature, 2018.(IF 40.13)