Anti-cFBPase - Cytosolic fructose-1,6-bisphosphatase (cytoplasm marker in photosynthetic tissues)
Cat# AS04043
Size : 50ul
Brand : Agrisera
Anti-cFBPase | Cytosolic fructose-1,6-bisphosphatase (cytoplasm marker in photosynthetic tissues)
AS04 043 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Brassica napus, Macroptilium atropurpureum, Nicotiana benthamiana, Pinus silvestris, Pinus yunanniensis, Oryza sativa, Petunia hybrida cv. Mitchell, Solanum tuberosum, Zea mays | cellular [compartment marker] of cytoplasm
- LI BAIYING | 2020-02-05This cFBAPase antibody is a very good one for western blot and shows specific and strong signals as a cytoplasm marker for Arabidopsis.Xiangfeng Wang | 2012-06-07This antibody shows very specific and strong signals for western blot using Arabidopsis materials.| 2011-12-07This antiserum is very specific and has a high titre.
| Immunogen: | Overexpressed cytosolic fructose 1,6 bisphosphatase (cFBPase) derived from the sequence from Arabidopsis thaliana cFBPase UniProt: Q9MA79, TAIR: AT1G43670 |
| Host: | Rabbit |
| Clonality: | Polyclonal |
| Purity: | Serum |
| Format: | Lyophilized |
| Quantity: | 50 µl |
| Reconstitution: | For reconstitution add 50 µl of sterile water |
| Storage: | Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. |
| Tested applications: | Immunolocalization (IL), Western blot (WB) |
| Recommended dilution: | 1: 500 (IL), 1 : 5 000 (WB) |
| Expected | apparent MW: | 45 | 37 kDa (Arabidopsis thaliana) |
| Confirmed reactivity: | Arabidopsis thaliana, Brassica napus, Macroptilium atropurpureum, Nicotiana benthamiana, Pinus silvestris, Pinus yunanniensis, Oryza sativa, Petunia hybrida cv. Mitchell, Solanum tuberosum, Zea mays |
| Predicted reactivity: | Capsella rubella, Pisum sativum, Ricinus communis, Glycine max, Phaseolus vulgaris, Sesamum indicum, Spinacia oleracea, Populus trichocarpa, Vitis vinifera Species of your interest not listed? Contact us |
| Not reactive in: | Chlamydomonas reinhardtii |
Application example 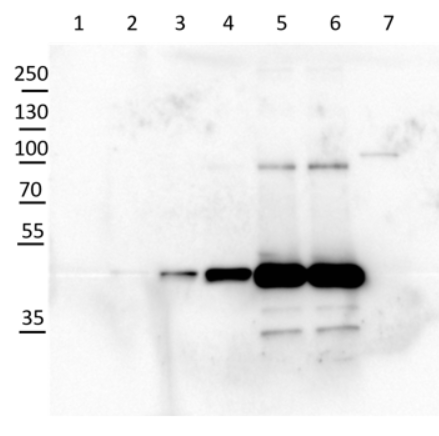 10 µg of Arabidopsis thaliana Col-0 WT chloroplast total protein (1), 10µg Col-0 WT chloroplast stroma protein (2), Col-0 WT total leaf sample 1:10 dilution (3), Col-0 WT total leaf sample 1:2 dilution (4), Col-0 WT total leaf sample undiluted (5), Col-0 WT total leaf sample undiluted (6) and recombinat plastidial FBPase 0.05 µg, expressed in E.coli with no cTP present in the sequence (7), extracted with 2x Laemmli buffer and denatured at 95°C for 5 min. were separated on 10% SDS-PAGE and blotted to Millipore Immobilon-P membrain (carried out at 100 V for 90 min at 4°C in blotting buffer (25 mM Tris-HCl, 192 mM glycine, 10 % [v/v] methanol). Blots were blocked with blocking solution ( TBST buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.05 % [v/v] Tween-20) supplemented with 5 % [w/v] milk powder) for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 5 000 for 15h (over night) at 4°C with agitation in TBST. The antibody solution was decanted and the blot was washed 6 times for 10 min in TBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:20 000 in for 1h at RT with agitation. The blot was washed as above and ChemiGlow West Chemiluminescence substrate was used for development according to the manufacturer’s instructions and imaged using the ChemiDoc imaging system (Biorad, Cressier, France). Exposure time was 15 seconds. Courtesy of Zanella Martina, ETH Zürich, Switzerland 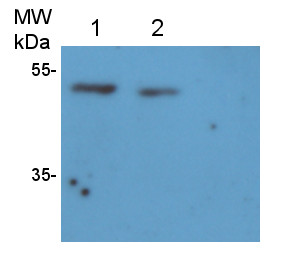 2 µg of total protein from Arabidopsis thaliana roots crude extract (1), supernatant after 10 0000 g centrifugation (2), extracted with ice-cold extraction buffer [50 mM Tris-HCl pH 7.5, 0.33 M Sucrose, 5 mM EDTA, 1x proteinase inhibitor] and denatured gradually with [50 mM Tris (pH 6.8), 10% Glycerol, 2% SDS, 2.5 M Urea, 0.005% Bromophenol Blue] at first with 55°C for 15 min followed by 95°C for 5 min. Proteins were separated on 12 % SDS-PAGE. Proteins were blotted 1h to PVDF using semi-dry transfer. Blots were blocked with 5 % nonfat milk + 0.1 % BSA for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 35 000 for ON/4°C with agitation in TBS-T. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase) diluted to 1:1 000 in for 2h at RT with agitation. The blot was washed as above and developed for 5 min withchemiluminescent detection reagent. Exposure time was 20 min. Courtesy of Dr. Joanna Jeleńska and Dequantarius Speed, University of Chicago, USA | |
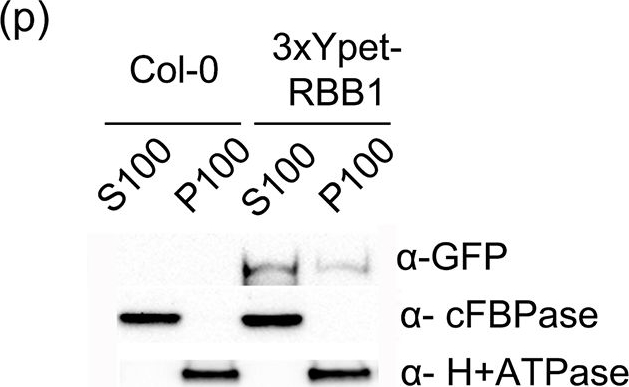
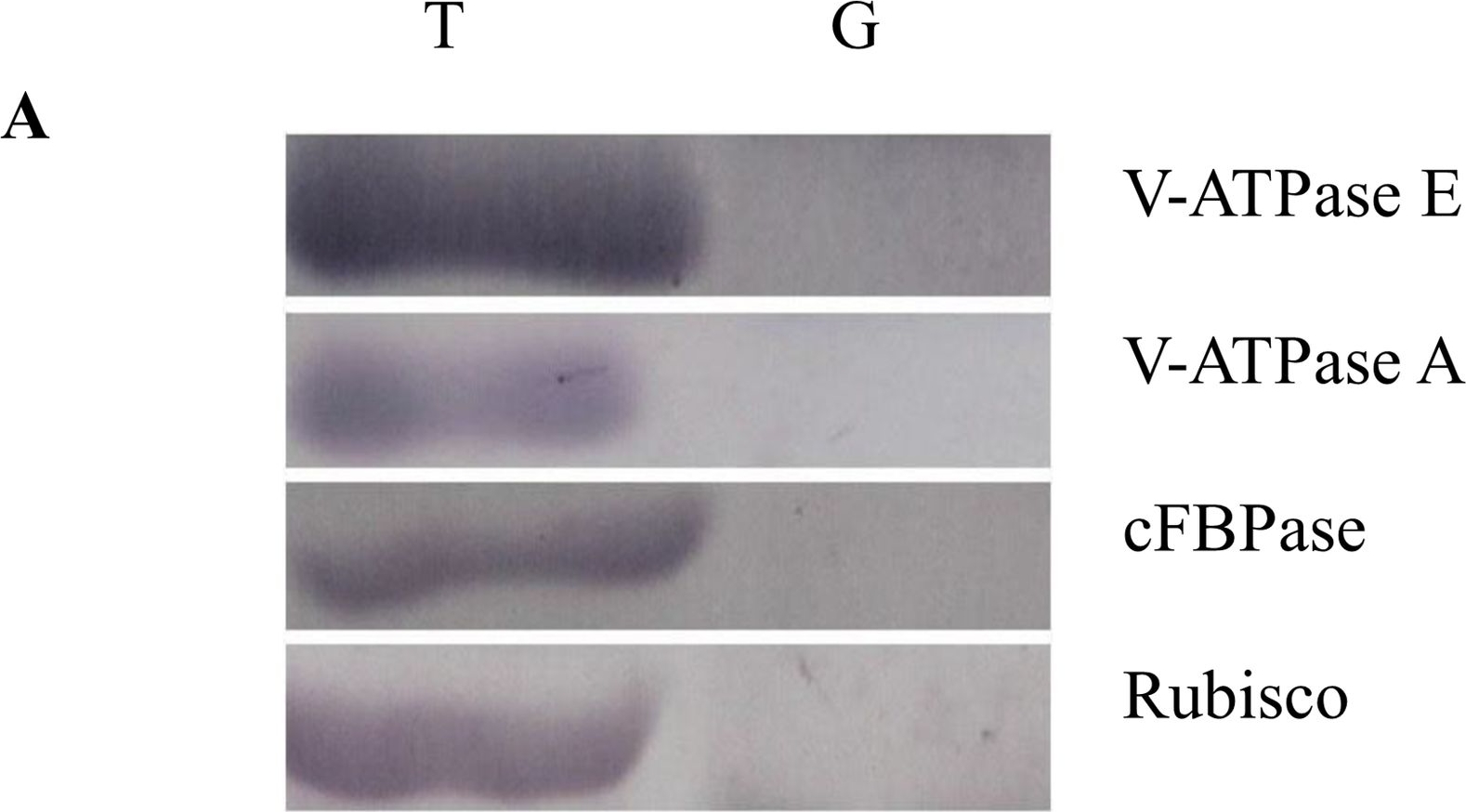
| Application examples: | Reactant: Arabidopsis thaliana (Thale cress) Application: Western Blotting Pudmed ID: 25915922 Journal: PLoS One Figure Number: 6p Published Date: 2015-04-29 First Author: Han, S. W., Alonso, J. M., et al. Impact Factor: 2.942 Open Publication3xYpet-RBB1 is a cytoplasmic protein that associates with the tonoplast.(a-c) A 3xYpet-RBB1 fusion complements the bulb phenotype of rbb1-2. Four-day-old seedlings from rbb1-2 (a) or rbb1-2 3xYpet-RBB1 (b, c) were stained with Lysotracker Red (magenta, a, b) to label the vacuole. The signal from 3xYpet-RBB1 (c) for the complemented plant is shown. (d-f) 3xYpet-RBB1 does not co-localize with FM4-64. Images show the cotyledon of 7-d-old seedlings expressing 3xYpet-RBB1 (d) stained with 5 ?M FM4-64 (e) and the merged image (f). (g-i) 3xYpet-RBB1 and RFP-SYP22 do not co-localize. Cotyledons of 7-d-old seedlings expressing 3xYpet-RBB1 (g) and RFP-SYP22 (h) were imaged. Merged image (i) is shown. (j-l) 3xYpet-RBB1 and Lysotracker Red do not co-localize in rosette leaves. Seedlings from the 3xYpet-RBB1 line were stained with Lysotracker Red. Signal from 3xYpet-RBB1 (j), Lysotracker Red (k) and the merged image (l) are shown. Arrowheads indicate the localization of 3xYpet-RBB1 at the tip of elongating trans-vacuolar strands. (m-o) GFP molecules can also label the tips of transvacuolar strands. Seedlings from a 35::GFP marker line were stained with Lysotracker Red. The GFP signal (m), Lysotracker Red (n) and the merged image (o) are shown. Arrowheads indicate the tip of TVS. All scale bars = 20 ?m. (p) 3xYpet-RBB1 accumulates in the soluble (S100) and membrane pellet (P100) fractions from whole seedlings. Immunoblot of 3xYpet-RBB1 soluble (S100) and membrane (P100) fractions from Col-0 and 3xYpet-RBB1 transgenic plants using antibodies against GFP (?-GFP), cFBPase (?-cFBPase, control for soluble fraction), and Plasma Membrane H+ATPase (?-H+ATPase, control for membrane fraction). Reactant: Oryza sativa (Asian rice) Application: Western Blotting Pudmed ID: 27992503 Journal: PLoS One Figure Number: 4A Published Date: 2016-12-20 First Author: Xing, S., Meng, X., et al. Impact Factor: 2.942 Open PublicationWestern blot image of the starch granule proteome. Same amount of proteins (25 ?g per lane) were loaded.A: Western blot images with different antibodies. The anti-bodies used were for V-ATPase E, V-ATPase A, Anti-Rubisco, cFBPase (Agrisera, Sweden). T: Total proteins; G: Starch granule proteins. B: Western blot image of protein acetylation of endosperm and starch granule proteins. Antibodies for acetylated Lysine (ImmuneChem) were used for Western blots. The source of proteins is indicated on the top of the lane. M: Protein marker; T: Total proteins extracted from endosperm; G: Proteins extracted from starch granules. |
| Additional information: | Kinetic and allosteric properties of the plant cytosolic FBPase are remarkably similar to the mammalian and yeast FBPase, but differ greatly from those of the chloroplastic FBPase. The antibody could detect FBPase from the human COS-7 cell line transfected with FBP1 expressing vector. |
| Additional information (application): | This antibody does not react with chloroplastic form of FBPase. |
| Background: | Fructose-1,6 bisphosphatase (FBPase) (EC=3.1.311) is one of the regulatory enzymes in the sucrose biosynthetic pathway. In non-photosynthetic tissues, it regulates the rate of gluconeogenesis. In photosynthetic tissues, two FBPase isozymes (chloroplastic and cytosolic) play key roles in carbon assimilation and metabolism. In photosynthetic tissues cFBPase (cytosolic fructose 1,6 bisphosphatase) converts triose phosphates from the chloroplast to sucrose during light hours. Alternative name: D-fructose-1,6-bisphosphate 1-phosphohydrolase |
| Selected references: | Ji et al (2023) The thioesterase APT1 is a bidirectional-adjustment redox sensor. Nat Commun. 2023 May 17;14(1):2807. doi: 10.1038/s41467-023-38464-y. Singh, Muthamilarasan, Prasad (2022). SiHSFA2e regulated expression of SisHSP21.9 maintains chloroplast proteome integrity under high temperature stress. Cell Mol Life Sci. 2022;79(11):580. Published 2022 Nov 3. doi:10.1007/s00018-022-04611-12 Cui, Liu, Li, et al. (2022) The cellulose--lignin balance affects the twisted growth of Yunnan pine trunk. Authorea. October 10, 2022. DOI: 10.22541/au.166538021.18232197/v2 Wang et al. (2022), Arabidopsis Ubiquitin-Conjugating Enzymes UBC4, UBC5, and UBC6 Have Major Functions in Sugar Metabolism and Leaf Senescence, Int. J. Mol. Sci. 2022, 23(19), 11143; https://doi.org/10.3390/ijms231911143 He, Gao, Luo, et al. (2022) VAMP724 and VAMP726 are involved in autophagosome formation in Arabidopsis thaliana [published online ahead of print, 2022 Oct 13]. Autophagy. 2022;1-18. doi:10.1080/15548627.2022.2127240 Lim et al (2022). Arabidopsis guard cell chloroplasts import cytosolic ATP for starch turnover and stomatal opening. Nat Commun. 2022 Feb 3;13(1):652. doi: 10.1038/s41467-022-28263-2. PMID: 35115512; PMCID: PMC8814037. Sun et al. (2021) Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana. Autophagy. 2021 Oct 17:1-17. doi: 10.1080/15548627.2021.1976965. Epub ahead of print. PMID: 34657568. Qian et al. (2021) OsFes1C, a potential nucleotide exchange factor for OsBiP1, is involved in the ER and salt stress responses, Plant Physiology, 2021;, kiab263. Shahbaz and Pilon (2019). Conserved Cu-MicroRNAs in Arabidopsis thaliana Function in Copper Economy under Deficiency. Plants (Basel). 2019 May 29;8(6). pii: E141. doi: 10.3390/plants8060141. Patir-Nebioglu et al. (2019). Pyrophosphate modulates plant stress responses via SUMOylation. Elife. 2019 Feb 20;8. pii: e44213. doi: 10.7554/eLife.44213. Liu et al. (2019). IMPORTIN Beta4 mediates nuclear import of GRF-INTERACTING FACTORs to control ovule development in Arabidopsis. Plant Physiol. 2019 Jan 18. pii: pp.01135.2018. doi: 10.1104/pp.18.01135. Seguel et al. (2018). PROHIBITIN 3 forms complexes with ISOCHORISMATE SYNTHASE 1 to regulate stress-induced salicylic acid biosynthesis in Arabidopsis. Plant Physiol. Jan 2018. DOI:10.1104/pp.17.00941 Lynch et al. (2017). Multifaceted plant responses to circumvent Phe hyperaccumulation by downregulation of flux through the shikimate pathway and by vacuolar Phe sequestration. Plant J. 2017 Dec;92(5):939-950. doi: 10.1111/tpj.13730. Duan et al. (2017). A Lipid-Anchored NAC Transcription Factor Is Translocated into the Nucleus and Activates Glyoxalase I Expression during Drought Stress. Plant Cell. 2017 Jul;29(7):1748-1772. doi: 10.1105/tpc.17.00044. (Nicotiana benthamiana) Steffens et al. (2017). Physical, Functional and Genetic Interactions between the BEACH Domain Protein SPIRRIG and LIP5 and SKD1 and Its Role in Endosomal Trafficking to the Vacuole in Arabidopsis. Front Plant Sci. 2017 Nov 20;8:1969. doi: 10.3389/fpls.2017.01969. Xing et al. (2016). Proteome Profile of Starch Granules Purified from Rice (Oryza sativa) Endosperm. PLoS One. 2016 Dec 19;11(12):e0168467. doi: 10.1371/journal.pone.0168467. LaMontagne et al. (2016). Isolation of Microsomal Membrane Proteins from Arabidopsis thaliana. Curr. Protoc. Plant Biol. 1:217-234. doi: 10.1002/cppb.20020. Ma et al. (2016). Phosphatidylserine Synthase Controls Cell Elongation Especially in the Uppermost Internode in Rice by Regulation of Exocytosis. PLoS One. 2016 Apr 7;11(4):e0153119. doi: 10.1371/journal.pone.0153119. eCollection 2016. de Michele et al. (2016). Free-Flow Electrophoresis of Plasma Membrane Vesicles Enriched by Two-Phase Partitioning Enhances the Quality of the Proteome from Arabidopsis Seedlings. J Proteome Res. 2016 Mar 4;15(3):900-13. doi: 10.1021/acs.jproteome.5b00876. Epub 2016 Feb 4. |
Protocols to work with plant and algal protein extracts