Anti-ARF1 - ADP-ribosylation factor 1
Cat# AS08325
Size : 50ul
Brand : Agrisera
Anti-ARF1 | ADP-ribosylation factor 1
AS08 325 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, C.reinhardtii, Elaeis sp. , L. longiflorum, M. truncatula, N. tabacum, O. sativa, P. hybrida cv Mitchell, P. patens, S. tuberosum | Cellular [compartment marker] of Golgi in immunolocalization and COP1 in Western blot
- Sukwon Choi | 2024-03-25I performed WB after Sucrose gradient fractionation. Anti-ARF1 was diluted 1:3,000 and Anti rabbit was diluted 1:10,000. Antibody works well.Tim Tranbarger | 2014-03-18We used the Arf1 antibody to examine the golgi position during the abscission process in the abscission zone of oil palm fruit and they worked very well for this monocot species and gave very interesting information. I plan to order and use other plant antibodies from Agrisera.Gerhard Obermeyer | 2009-11-18The antibody worked well in Western blots and gave a main signal at 21 kDa in a microsomal fraction prepared from Lilium pollen grains. Anti-Arf1 was diluted 1:1,000 and an anti-rabbit IgG AP-conjugated (1:5,000) was used for detection.Antonio Castillo-Flores | 2009-03-03I tried this Arabidopsis antibody with Chlamydomonas cell extract on a Western (1:1000 ECL, 1:20000 anti-Rabbit, both incubated for 1h in a rotator at room temperature).The abs worked nice. It gave a unique and clear band with a MW around 19 kD (The MW of Arf1 in Arabidopsis is 21 kD).I grade the abs with 5/5. However, I give a grade of 3/5 to the concentration of the abs in the 200 ul serum.
| Immunogen: | Recombinant GST fusion of full length of Arabidopsis thaliana ARF1 (P36397, AT2G47170) |
| Host: | Rabbit |
| Clonality: | Polyclonal |
| Purity: | Serum |
| Format: | Lyophilized |
| Quantity: | 50 µl |
| Reconstitution: | For reconstitution add 50 µl of sterile water |
| Storage: | Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. |
| Tested applications: | Immunofluorescence (IF), Immunogold (IG), Western blot (WB) |
| Recommended dilution: | 1 : 1000 (IF), 1 : 100 (IG), 1 : 1000 (WB) |
| Expected | apparent MW: | 21 kDa (Arabidopsis thaliana) |
| Confirmed reactivity: | Actinidia chinensis, Arabidopsis thaliana, Chlamydomonas reinhardtii, Elaeis sp. , Lilium longiflorum, Medicago truncatula, Nicotiana tabacum, Oryza sativa, Petunia hybrida cv Mitchell, Physcomitrium patens, Solanum tuberosum |
| Predicted reactivity: | Brassica juncea, Brassica napus, Capsella rubella, Capsicum annum, Cucumis sp., Daucus carota, Elaeis guineensis, Glycine max, Helleborus orientalis, Hordeum vulgare, Medicago truncatula, Nannochloropsis gaditana, Nicotiana benthamina, Ostreococcus tauri, Populus trichocarpa, Triticum aestivum, Zea mays Species of your interest not listed? Contact us |
| Not reactive in: | Centella asiatica, Microsporidia sp. |
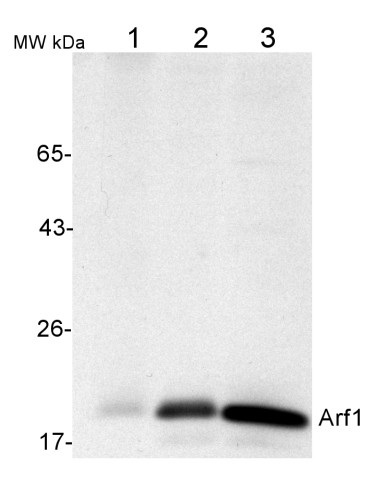
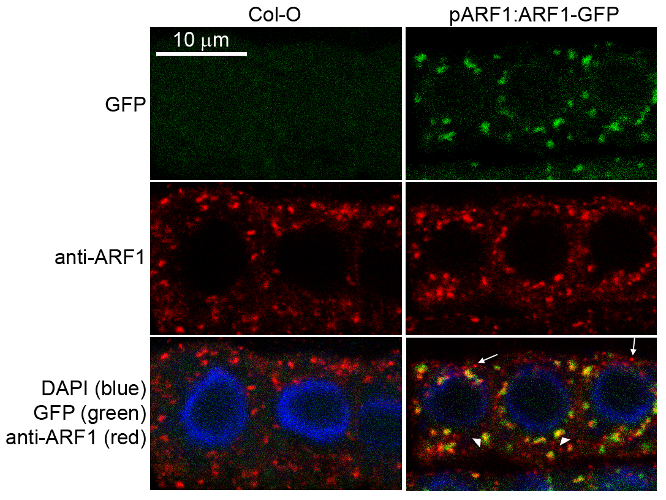
| Application example 50 µg of total protein from (1) Nicotiana tabacum protoplast total protein, (2) Arabidopsis thaliana protoplast soluble protein, (3) Arabidopsis thaliana protoplast total protein were separated on 10 % SDS-PAGE and blotted 2h to nitrocellulose (Semi-dry, 200mA). Filters were blocked over night with 5% low-fat milk powder in TBS and probed with anti-Arf1 antibodies (AS08 325, 1:1000, 1h) and secondary anti-rabbit (1:20000, 1 h) antibody (HRP) in TBS-Tween (recommended secondary antibody AS09 602). Signal was detected with chemiluminescence detection reagent and exposure time for this image was 1 minute. Protoplasts were extracted in 50mM Tris, 10 mM EDTA and Triton X100, 0.02%. Immunofluorescence Specificity testing of rabbit anti-ARF1 serum. Immunofluorescence labelling of rabbit anti-ARF1 antibody (red) in 5-day-old root epidermal cells of the Arabidopsis thaliana ecotype Columbia-0 (WT) or seedlings expressing the ADP-RIBOSYLATION FACTOR 1 (AtARFA1c; accession At2g47170) fused to EGFP (green) (Xu, J. and Scheres, B. 2005. Plant Cell 17, 525-536). The rabbit anti-ARF1 antibody was diluted 1:1000 and the secondary antibody, donkey anti-rabbit CY5-coupled (Jackson ImmunoResearch) was diluted 1:300. The nuclei were stained with DAPI (blue). Note the co-labelling of ARF1-GFP with the anti-ARF1 antibody (arrowheads) and the additional labelling (potentially of other ARF1 variants) by the anti-ARF1 antibody (arrows). The antibody staining permeability was limited to the 1-2 outermost layers of the whole-mounted root tips. Courtesy of Dr. Anna Gustavsson and Dr. Markus Grebe | |
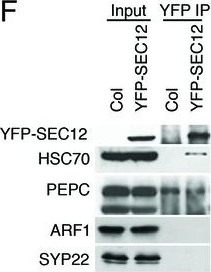
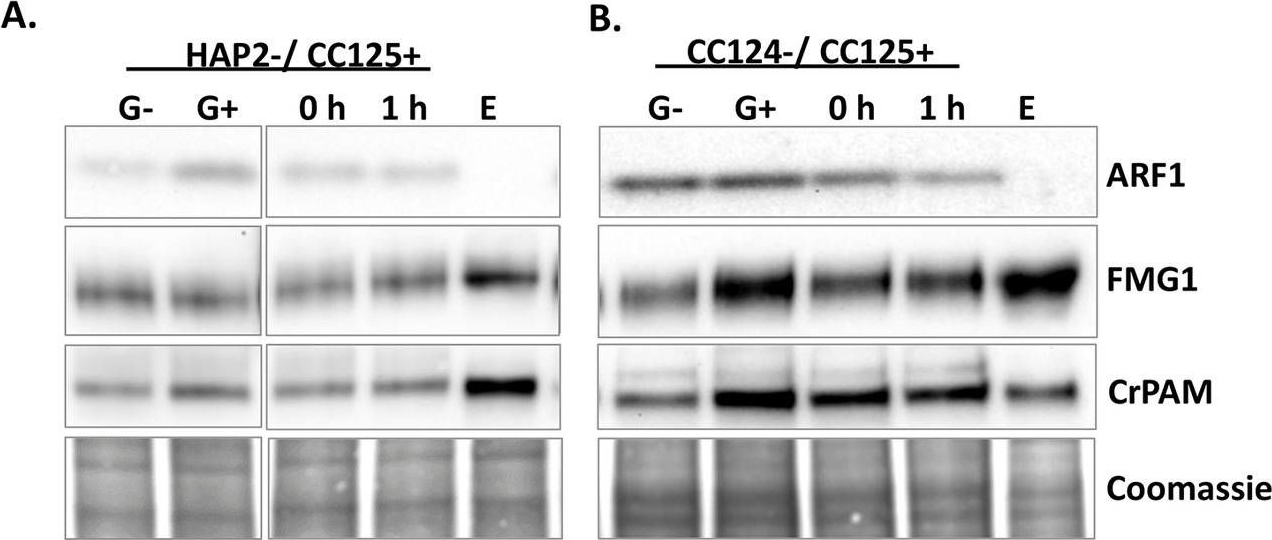
| Application examples: | Reactant: Arabidopsis thaliana (Thale cress) Application: Western Blotting Pudmed ID: 27938667 Journal: Elife Figure Number: 1F Published Date: 2016-12-12 First Author: Li, S., Le, B., et al. Impact Factor: 7.448 Open PublicationAnalyses of sRNA-seq libraries.(A) Hierarchical clustering analysis showing the degree of similarity among the sRNA-seq libraries. Sample-to-sample distances were calculated based on log-transformed normalized read counts. The biological replicates of each sample type were highly reproducible. T: total extract; M: microsome; TP: total polysome; MBP: membrane-bound polysome; T27: sRNA-seq from total extracts in ago1-27; M27: sRNA-seq from microsomes in ago1-27. The other samples were from wild type. The numbers after the underscore represent biological replicates. (B) Size distribution of sRNAs in the indicated samples. T: total extract; M: microsome; ER-IP: ER immunoprecipitation. Note that normalization was performed against total r/t/snRNA-depleted reads. (C) The composition of sRNAs in each of three size classes. Each column represents a biological replicate. Note that normalization was performed against total r/t/snoRNA-depleted reads. (D) Western blotting using anti-GFP antibodies to show that YFP-SEC12 was in the microsomal fraction. (E) Topology mapping showing that the YFP tag of the ER transmembrane protein YFP-SEC12 was on the cytosolic side. (F) Western blotting showing that ER immunoprecipitation (using anti-GFP antibodies with the YFP-SEC12 transgenic line) contained two ER markers (YFP-SEC12 and HSC70) but not markers for Golgi (ARF1) or endosomes (SYP22). PEPC is a soluble protein. Col, wild type (without the YFP-SEC12 transgene). (G) A scatter plot showing the depletion of Pol IV-dependent siRNAs (P4siRNAs) from the microsomal fraction. Each dot represents a 100 bp window that generates P4siRNAs. Note that normalization was performed using rRNA fragments as internal controls. (H–K) Wild type and nrpd1-1 sRNA-seq samples that differ greatly in sRNA compositions were used as a proof-of-concept for the rRNA fragment-based normalization method. (H–I) Normalization using total r/tRNA-depleted, genome-matched reads led to an exaggeration of the 21-nt peak (H) and an apparent increase in the abundance of miRNAs (I) in nrpd1-1, which lacks most 24-nt siRNAs. (J–K) Normalization using rRNA fragments resulted in similar abundance of 21-nt siRNAs (J) and miRNAs (K) between wild type and nrpd1-1.DOI:http://dx.doi.org/10.7554/eLife.22750.003 Reactant: Chlamydomonas reinhardtii (Green Alga) Application: Western Blotting Pudmed ID: 31809498 Journal: PLoS Biol Figure Number: 7A,B Published Date: 2019-12-01 First Author: Luxmi, R., Kumar, D., et al. Impact Factor: 7.279 Open PublicationRelease of CrPAM in ciliary ectosomes is developmentally regulated.A. HAP2 minus and CC125 plus gametes, mating cells (0- or 1-hour), and ectosome-rich pellets prepared from the 1-hour media were analyzed. Equal amounts of protein (30 ?g) were fractionated and subjected to immunoblot analysis for ARF1, FMG1, and CrPAM. Quantification of FMG1 and CrPAM protein levels is shown in the lower panels; results are the average of two independent experiments—error bars indicate the range. B. Cell lysates and ectosomes prepared from wild-type CC124?/CC125+ gametes were analyzed as described for panel A. Results are the average of six independent experiments; error bars indicate ± SEM. Asterisks indicate a statistically significant difference between two groups (*P < 0.01). C. and D. Immunoblot analysis of cells and ectosomes harvested from vegetative CC124? (C) and CC125+ (D) cells; quantification of FMG1 and CrPAM levels is shown below. Results are the average of three experiments; error bars indicate ± SEM. CrPAM and FMG1 levels in vegetative ectosomes differed significantly from levels in cells (**P = 0.0065, ***P < 0.0001). E. CrPHM and CrPAL activities were assayed in cells and ectosomes released by vegetative CC124? and CC125+ cells, mating HAP2?/CC125+ cells, and mating CC124?/CC125+ cells. Both activities were significantly higher in ectosomes released by mating gametes (*P < 0.01, **P < 0.001, ***P < 0.0001; one-way ANOVAs). F. Immunogold-electron microscopy negative stain image showing localization of CrPAM on mating ectosomes with antibody against CrPAM luminal domain. Negative control, ectosomes incubated with gold-tagged secondary antibody alone. The underlying numeric data for this figure can be found in S1 Data. ARF1, ADP ribosylation factor 1; FMG1, flagellar membrane glycoprotein 1. |
| Additional information: | Cellular [compartment marker] of Golgi in immunolocalization and COP1 in western blot |
| Additional information (application): | References describing immunolocalization (IF) and (IG) studies: Pimpl et al (2000). In Situ Localization and in Vitro Induction of Plant COPI-Coated Vesicles. Plant Cell. 2000 Nov;12(11):2219-36. Ritzenthaler et al. (2002). Reevaluation of the Effects of Brefeldin A on Plant Cells Using Tobacco Bright Yellow 2 Cells Expressing Golgi-Targeted Green Fluorescent Protein and COPI Antisera. Plant Cell. 2002 Jan;14(1):237-61. |
| Background: | The ARF1 protein is localized to the Golgi apparatus and has a central role in intra-Golgi transport. It is a small GTPase that undergoes a GDP/GTP nucleotide exchange cycle and it is an important regulator of cellular trafficking. |
| Selected references: | Mohr et al. (2024). Characterization of the small Arabidopsis thaliana GTPase and ADP-ribosylation factor-like 2 protein TITAN. J Cell Sci . 2024 Aug 14;137(15):jcs262315. doi: 10.1242/jcs.262315. (immunofluorescence) Suanno et al. (2023) Small extracellular vesicles released from germinated kiwi pollen (pollensomes) present characteristics similar to mammalian exosomes and carry a plant homolog of ALIX, Front. Plant Sci., 25 January 2023, Sec. Plant Membrane Traffic and Transport, Volume 14 - 2023. Chien et al. (2022) Phosphate transporter PHT1;1 is a key determinant of phosphorus acquisition in Arabidopsis natural accessions. Plant Physiol. 2022 Aug 29;190(1):682-697. doi: 10.1093/plphys/kiac250. PMID: 35639954; PMCID: PMC9434223. (Immunogold). Brumm, Singh, Kriechbaum, et al. (2022) N-terminal domain of ARF-GEF GNOM prevents heterodimerization with functionally divergent GNL1 in Arabidopsis. Plant J. 2022;112(3):772-785. doi:10.1111/tpj.15979. Farago et al. (2022) Small paraquat resistance proteins modulate paraquat and ABA responses and confer drought tolerance to overexpressing Arabidopsis plants. Plant Cell Environ. 2022 Jul;45(7):1985-2003. doi: 10.1111/pce.14338. Epub 2022 Apr 29. PMID: 35486392; PMCID: PMC9324991. Narasimhan et al. (2021) Systematic analysis of specific and nonspecific auxin effects on endocytosis and trafficking. Plant Physiol. 2021 Mar 18:kiab134. doi: 10.1093/plphys/kiab134. Epub ahead of print. PMID: 33734402. Hurny et al. (2020). SYNERGISTIC ON AUXIN AND CYTOKININ 1 Positively Regulates Growth and Attenuates Soil Pathogen Resistance. Nat Commun. 2020 May 1;11(1):2170. doi: 10.1038/s41467-020-15895-5. (immunolocalization) Kuang et al. (2019). Quantitative Proteome Analysis Reveals Changes in the Protein Landscape During Grape Berry Development With a Focus on Vacuolar Transport Proteins. Front Plant Sci. 2019 May 15;10:641. doi: 10.3389/fpls.2019.00641. eCollection 2019. Singh et al. (2018). A single class of ARF GTPase activated by several pathway-specific ARF-GEFs regulates essential membrane traffic in Arabidopsis. PLoS Genet. 2018 Nov 15;14(11):e1007795. doi: 10.1371/journal.pgen.1007795. Lang et al. (2011).Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011 Feb;30(2):205-15.doi: 10.1007/s00299-010-0935-4. Gonzaga Heredia-Martinez et al. (2018). Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Physiol, Sept. 2018. Lynch et al. (2017). Multifaceted plant responses to circumvent Phe hyperaccumulation by downregulation of flux through the shikimate pathway and by vacuolar Phe sequestration. Plant J. 2017 Dec;92(5):939-950. doi: 10.1111/tpj.13730. Vincent et al. (2017). A genome-scale analysis of mRNAs targeting to plant mitochondria: upstream AUGs in 5' untranslated regions reduce mitochondrial association. Plant J. 2017 Dec;92(6):1132-1142. doi: 10.1111/tpj.13749. Ma et al. (2016). Phosphatidylserine Synthase Controls Cell Elongation Especially in the Uppermost Internode in Rice by Regulation of Exocytosis. PLoS One. 2016 Apr 7;11(4):e0153119. doi: 10.1371/journal.pone.0153119. eCollection 2016. Yuzbasıoglu et al. (2016). Functional specialization of Arf paralogs in nodules of model legume, Medicago truncatula. Plant Growth Regul. DOI: 10.1007/s10725-016-0227-2. Marais et al. (2015). The Qb-SNARE Memb11 interacts specifically with Arf1 in the Golgi apparatus of Arabidopsis thaliana. J Exp Bot. 2015 Jul 24. pii: erv373. Wang et al. (2015). UDP-D-galactose synthesis by UDP-glucose 4-epimerase 4 is required for organization of the trans-Golgi network/early endosome in Arabidopsis thaliana root epidermal cells. J. Plant Res. 2015 May 27. (immunogold application). |
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela